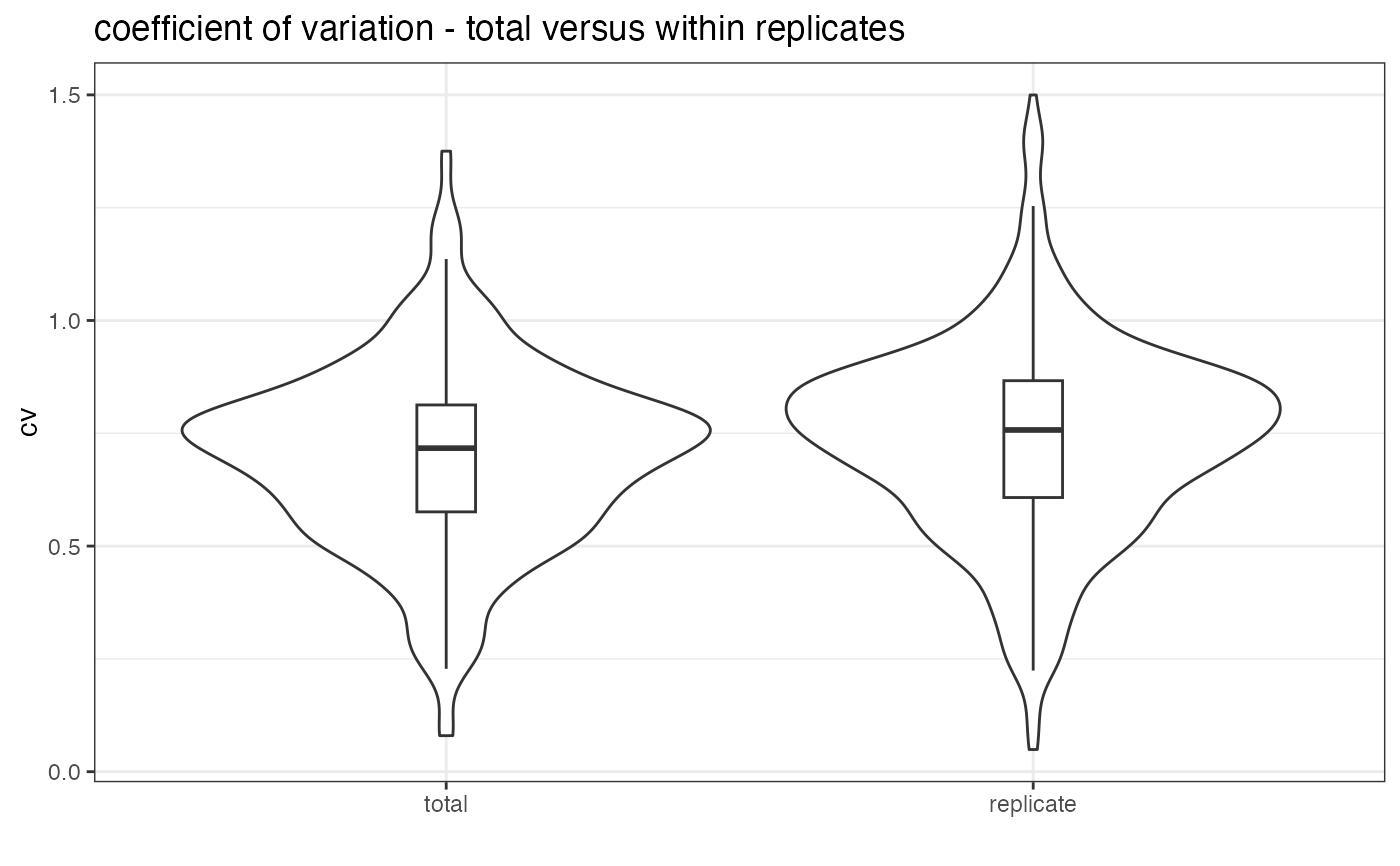
Plots the total variation versus variation within replicates
Source:R/plot_variation_vs_total.R
plot_variation_vs_total.RdThis function plots the total variation and the variation within replicates for a given value. If decoys are present these are removed before plotting.
plot_variation_vs_total(
data,
column.values = "Intensity",
comparison1 = transition_group_id ~ BioReplicate + Condition,
comparison2 = transition_group_id + Condition ~ BioReplicate,
fun_aggregate = NULL,
label = FALSE,
title = "coefficient of variation - total versus within replicates",
boxplot = TRUE,
...
)Arguments
- data
Data table that is produced by the OpenSWATH/pyProphet workflow.
- column.values
Indicates the columns for which the variation is assessed. This can be the Intensity or Signal, but also the retention time.
- comparison1
The comparison for assessing the total variability. Default is to assess the variability per transition_group_id over the combination of Replicates and different Conditions.
- comparison2
The comparison for assessing the variability within the replicates. Default is to assess the variability per transition_group_id and Condition over the different Replicates.
- fun_aggregate
If depending on the comparison values have to be aggregated one needs to provide the function here. (I think this should be sum, yesno?)
- label
Option to print value of median cv.
- title
Title of plot. Default: "cv across conditions"
- boxplot
Logical. If boxplot should be plotted. Default: TRUE
- ...
Arguments passed through, currently unused.
Value
Plots in Rconsole a violin plot comparing the total variation with the variation within replicates. In addition it returns the data frame from which the plotting is done and a table with the calculated mean, median and mode of the cv for the total or replicate data.
Examples
{
data("OpenSWATH_data", package="SWATH2stats")
data("Study_design", package="SWATH2stats")
data <- sample_annotation(OpenSWATH_data, Study_design)
var_summary <- plot_variation_vs_total(data)
}