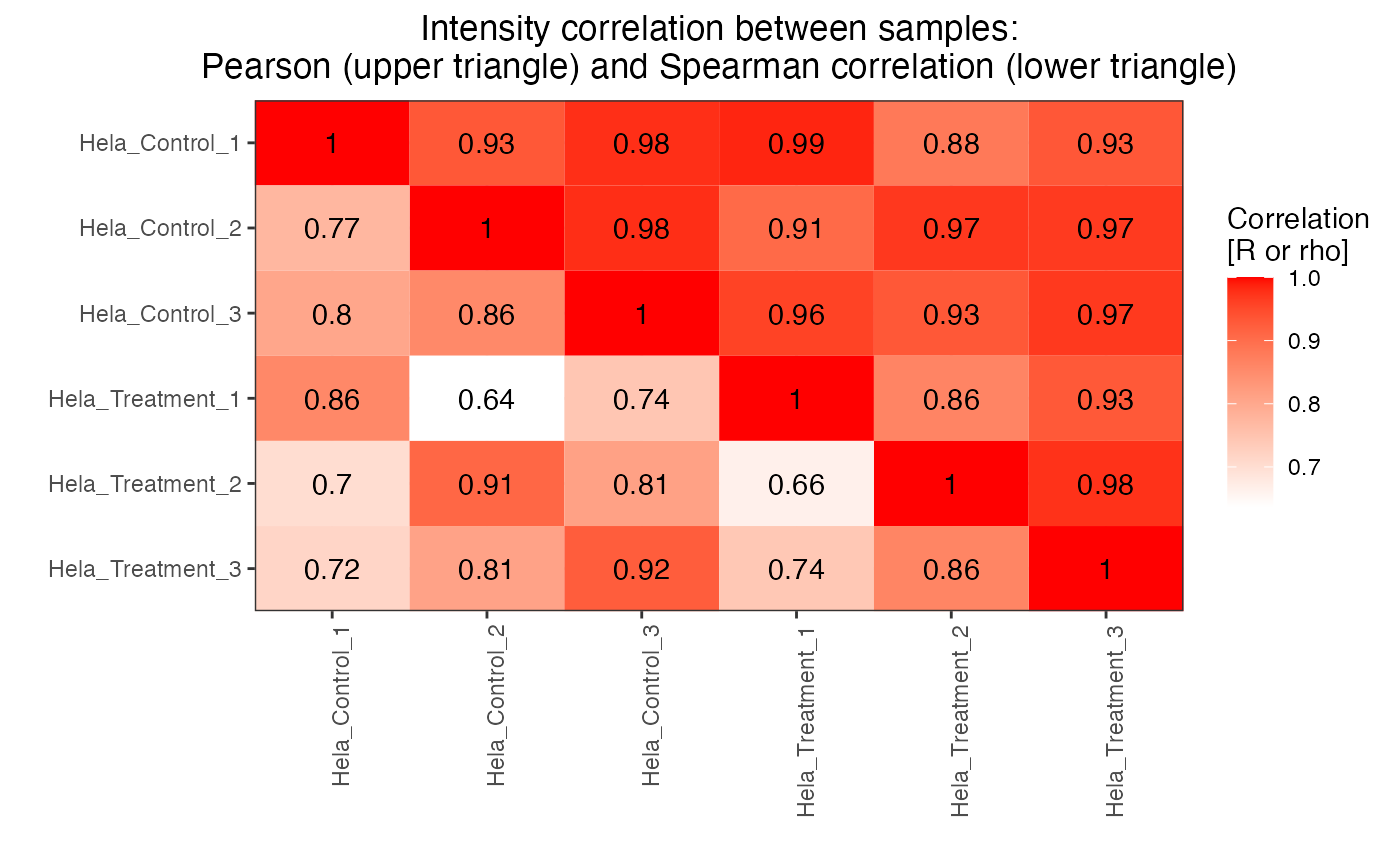
Plots the correlation between injections.
Source:R/plot_correlation_between_samples.R
plot_correlation_between_samples.RdThis function plots the Pearson's and Spearman correlation between samples. If decoys are present these are removed before plotting.
plot_correlation_between_samples(
data,
column_values = "Intensity",
comparison = transition_group_id ~ Condition + BioReplicate,
fun_aggregate = NULL,
label = TRUE,
...
)Arguments
- data
Data frame that is produced by the OpenSWATH/pyProphet workflow.
- column_values
Indicates the columns for which the correlation is assessed. This can be the Intensity or Signal, but also the retention time.
- comparison
The comparison for assessing the variability. Default is to assess the variability per transition_group_id over the different Condition and Replicates. Comparison is performed using the dcast() function of the reshape2 package.
- fun_aggregate
If for the comparison values have to be aggregated one needs to provide the function here.
- label
Option to print correlation value in the plot.
- ...
Further arguments passed to methods.
Value
Plots in Rconsole a correlation heatmap and returns the data frame used to do the plotting.
Examples
{
data("OpenSWATH_data", package="SWATH2stats")
data("Study_design", package="SWATH2stats")
data <- sample_annotation(OpenSWATH_data, Study_design)
information <- plot_correlation_between_samples(data)
}